At CBR, we continue to develop methods for synthetic biotechnology. With them, we break new ground or improve existing processes.
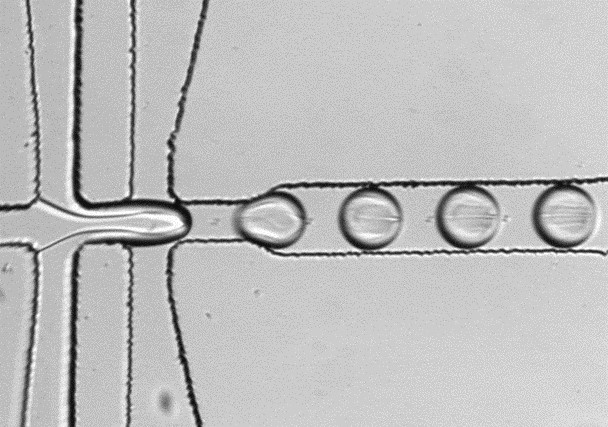
Our focus is on the development of biochemical assays for absorption-based ultra-high-throughput droplet microfluidics. These assays are used, for example, in enzyme engineering experiments to screen particularly large libraries. In addition, we combine droplet microfluidics with synthetic biology methods to identify new enzymes from metagenomes in high throughput.
Droplet microfluidics can miniaturise biochemical assays from the usual µL scale to the picolitre scale. Droplets are monodisperse droplets that can be automatically produced by thousands per second using microfluidic chips.
The individual droplets function as closed reaction chambers and can be used in a similar way to the individual wells in microtitre plates. Microfluidic chips equipped with detectors can, for example, read absorption signals from droplets. Based on these signals, individual droplets can be sifted in special sorting chips, with a throughput of up to 300 Hz, and specifically isolated for further analysis.
This considerably improves throughput and resource consumption compared to conventional microtitre plate assays.
Biological cell factories can sustainably produce a wide range of products such as fuels or pharmaceuticals. In this way, they support more environmentally friendly industrial processes and the necessary shift towards a bio-based economy. However, the development of these cell factories is currently time-consuming and labour-intensive because, among other things, attention must be paid to the integrity of the organism used. However, by transferring the reaction from inside the cell to the free solution, these limitations can be overcome. Therefore, the sufficient provision of proteins for synthetic enzyme cascades (in vitro) represents an important field of development.
The yeast Pichia pastoris is one of the most important heterologous expression systems in biotechnology. In addition to the secretion of proteins from the cell, post-translational modifications (PTM) such as glycosylation are also possible. Secreted proteins can subsequently be applied more cost-effectively on an industrial scale. Although P. pastoris is widely used for industrial applications and has already been intensively studied, an efficient strategy that allows the systematic identification of optimal expression and secretion conditions is lacking. Therefore, a modular toolbox of standardised genetic elements and different secretion signals was developed at the Department of Chemistry of Biogenic Resources (keyword synthetic biology). These elements can be randomly combined into large and diverse libraries in an uncomplicated way using hierarchical assembly. The identification of the best combination is carried out robotically in high-throughput screening (link). Using the developed method, new combinations could be identified that led to increased protein secretion. The flexible design of the platform provided here enables efficient optimisation in the high-throughput process and thus represents a valuable resource for future development projects.
With the know-how and equipment available at the department, a wide range of laboratory work can be automated and carried out with high throughput.
Established procedures are, for example:
- Screening of strain collections for polysaccharide producers
- Screening of enzyme libraries for activity, stability or substrate specificity
- extraction and purification of proteins, RNA and DNA
- absorptive and chromatographic separation methods in 96-well format (e.g. SEC or IMAC)
- enzymatic and chemical quantification methods, e.g. for glucose, pyruvate, proteins
- enzyme activity assays
- Enzyme kinetics
- Optimisation experiments, e.g. medium optimisation